Step 2: Geostatistical Hierarchical Clustering
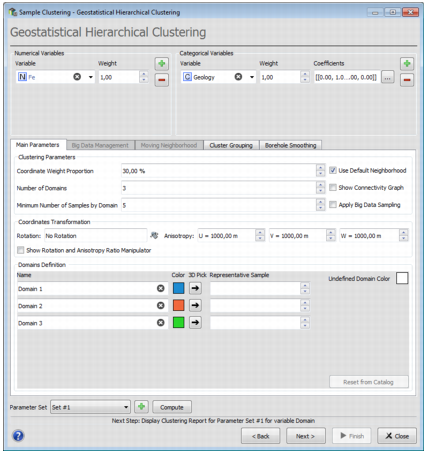
Variable and Weights
-
Numerical Variable:
Use
 and
and  to add or remove a variable. Select a float or an integer variable but not an alphanumerical variable. Once the variable is selected, enter a positive weight.
to add or remove a variable. Select a float or an integer variable but not an alphanumerical variable. Once the variable is selected, enter a positive weight. -
Categorical Variable:
Use
 and
and  to add or remove a variable. Select only categorical variables. Once the variable is selected, enter a positive weight.
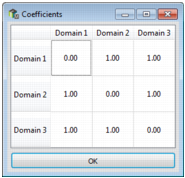
to add or remove a variable. Select only categorical variables. Once the variable is selected, enter a positive weight. Click on ... to edit the coefficients matrix that defines the dissimilarity between each possible pair of values. The coefficient on the diagonal must be equal to 0 and the matrix is symmetric.
Main Parameters
Clustering Parameters
-
Coordinate Weight Proportion: Set a value to the Coordinates Weight Proportion. This weight will be applied to the Euclidean distance (X, Y & Z) between two given samples.
To simplify, this weight is given in percent relatively to the sum of all the weights (Coordinates, Numerical and Categorical Variables).
For example, if a 50% weight is assigned to the Coordinates, a 3 weight is assigned to a Numerical variable and a 2 weight is assigned to a Categorical variable, the weight of the Coordinates is unchanged and equal to 50%, the weight of the numerical variable is 30% and the weight of the categorical variable is 20%.
As a consequence if the Coordinate Weight Proportion is set to 100%, all the other variables are ignored during the clustering operation and if the Coordinate Weight Proportion is set to 0, the coordinates are ignored during the clustering operation.
- Number of Domains: Enter the number of domains to be kept.
- Minimum Number of Samples by Domain: If a domain is made of less samples than the Minimum Number of Samples by Domain, the domain won’t be taken into account and its samples will be associated with the Undefined Domain.
- Use Default Neighborhood: Select this option to use a default moving neighborhood. Otherwise, uncheck this option and edit neighborhood characteristics in the Moving Neighborhood tab. It corresponds to a 8 splitted sectors neighborhood with an optimum/maximum number of samples per sector equal to 1 and an infinite size.
- Show Connectivity Graph: Check this option to display the sample network in the 3D View of the graphical output window after pressing Compute or Next.
- Apply Big Data Sampling: Activate this toggle if your data set is bigger than 1000 samples to make the clustering operation faster.
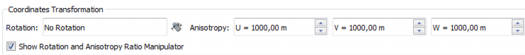
Coordinates Transformation
After the sample network has been established, euclidean distances will be computed to contribute to the dissimilarity matrix (using above weight). Those distances are computed using transformed coordinates applying the given Rotation and Anisotropy Ratio. This is useful when dealing with highly anisotropic sampling pattern to normalize the coordinates.
Select the Show Rotation and Anisotropy Ratio Manipulator option to have access to the interactive tool in the 3D View of the graphical output window.
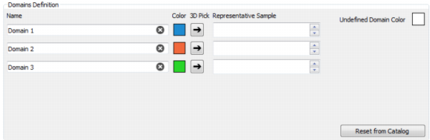
Domain Definition
For each domain:
- Domain Name can be edited in the corresponding field.
- Domain Color can be edited by clicking on the colored square.
-
If a run has already been applied you can pick a sample displayed in the 3D View of the graphical output window and press on the 3D Pick button
 to use this sample has a representative sample for a giving domain.
to use this sample has a representative sample for a giving domain.
The Undefined Domain gathers samples that could not be classified into the domains. The color of the undefined domain can be edited.
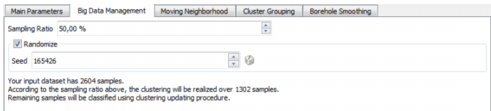
Big Data Management
Note: This option is available if the toggle Apply Big Data Sampling is activated in the Main Parameters tab.
The Big Data Management enables you to sample your data and to reduce the time of computation.
- Sampling Ratio: Specify a Sampling Ratio, on which the GHC method will be applied. The remaining samples will be classified using the SVM method (Support Vector Machines). A Sampling Ratio of 30% will drastically reduce the computation time with a small impact on the output quality.
- Randomize: Select this toggle to randomize the sampling. Click on the dice
 to generate automatically random seed numbers.
to generate automatically random seed numbers.
Moving Neighborhood
Note: This option is only available if the toggle Use Default Neighborhood is off in the Main Parameters tab. It is advised to use the default moving neighborhood.
The Samples are selected in the neighborhood only if they fall inside an ellipsoid centered on the target point. When a sample is selected, a connection is created with the target point. If the neighborhood search fails because of some criterion, the target point won’t be linked with any neighbors.
This ellipsoid is defined through its dimensions along its main axes and a possible rotation around the main axes of the study.
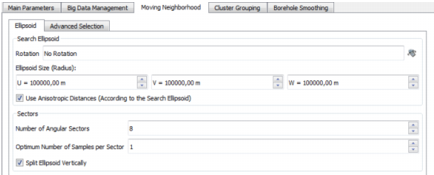
Ellipsoid Parameters
-
Search Ellipsoid
- Click Rotation to pop up the Rotation Definition dialog box and define a possible rotation of the Search Ellipsoid.
- Ellipsoid Size (Radius): Enter the ellipsoid dimensions along the three main axes (U, V, W) of the Search Ellipsoid (U, V, W standing for the rotated X, Y, Z).
-
The application first calculates the distances between each sample that is inside the Search Ellipsoid and the cell gravity center. Then it sorts the samples as a function of this calculated distance. The closest samples from the target point are preferentially selected. These distances can be calculated in two different ways:
- if the option Use Anisotropic Distances (According to the Search Ellipsoid) is not selected, the distances will be isotropic standard distances.
-
if the option Use Anisotropic Distances (According to the Search Ellipsoid) is selected, the distances will be anisotropic and calculated taking into account the Search Ellipsoid. For example, all the samples located on the boundary of the ellipsoid are considered to be at the same distance from the target point.
This is particularly useful when the sampling pattern is highly anisotropic or in 3D to take samples horizontally at a greater distance than vertically.
-
Sectors
- Number of Angular Sectors: The ellipsoid can be split in different angular sectors inside which the neighbors are grouped. You can increase the number of angular sectors in order to make sure that some samples are selected from different directions in the field. This is particularly useful when the sampling pattern is highly anisotropic.
- Optimum Number of Samples per Sector: The search for samples in the different sectors is a sequential process. The application scans each sector at a time until it has been assigned an Optimum Number of Samples, if possible, within the ellipsoid.
- Split Ellipsoid Vertically: Select this option to double the number of angular sectors.
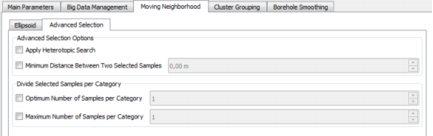
Advanced Selection Parameters
This tab groups advanced parameters used to refine the neighborhood.
-
Advanced Selection Options
-
Apply Heterotopic Search: When performing a multivariate neighborhood search, some variables may be undefined for some of the samples. This is called the heterotopic case. Conversely, the isotopic case indicates that all the variables are defined for all the samples.
- Leave this option clear if you wish to perform a standard neighborhood search. The Optimum Number of Samples will be reached for each sector although the selected information may not be convenient for the cokriging process. This is the case, for instance, when one of the variables is not sufficiently represented.
- Select this option if you wish to get more information from the variables which are defined on less samples. A preliminary step is added. The algorithm still looks for the nearest samples in each sector, but tries to get all the variables informed whatever the number of samples needed to fulfill this additional requirement. Each variable will be informed in at least one of the selected samples of each sector. After this step, the search goes up in a standard way.
- Sometimes, increasing the count of angular sectors is not always sufficient to stabilize the neighborhood. Select the option Minimum Distance Between Two Selected Samples if you wish to counterbalance any clustered configuration. As soon as one sample has been selected, it won’t be possible to select any other sample located at a smaller distance than the defined one.
-
-
Divide Selected Samples per Category
- Optimum Number of Samples per Borehole: If the optimum number of samples per borehole has not been reached, a second pass is performed to select the nearest samples that have been ignored during the first search.
- Maximum Number of Samples per Borehole: If the maximum number of samples on a borehole have been selected, it will not be possible to select any other sample from this borehole even if the optimum number of samples per sector has not been reached.
Note: The Optimum Number of Samples per Borehole should be lower than the Maximum Number of Samples per Borehole and lower than the Optimum Number of Samples per Sector.
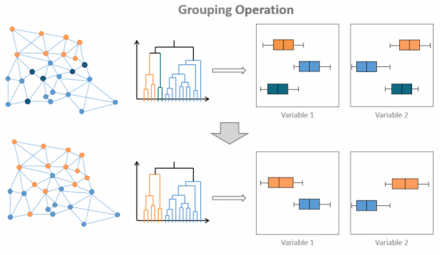
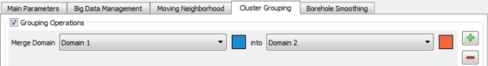
Cluster Grouping
This tab is dedicated to the Cluster Grouping post-processing.
It is meant to merge a domain into another one.
Use ![]() and
and ![]() to add and remove a rule.
to add and remove a rule.
Select two Domains. On this figure the Domain 1 will be merged into the Domain 2.
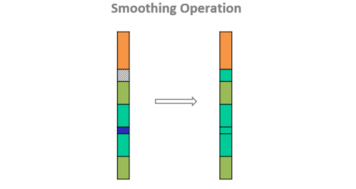
Borehole Smoothing
This tab is dedicated to the Borehole Smoothing post-processing.
It is meant to smooth the borehole by removing small intervals and to fill the gap interval.
-
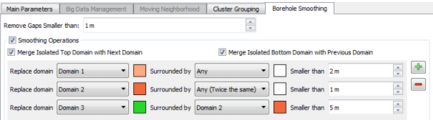
Enter a distance value to Remove Gaps Smaller than this given distance. The undefined sequence is replaced by the most likely domain using domain transition probabilities.
Note: To avoid the Remove Gaps operation set the parameter value to zero.
- Merge Isolated Top Domain with the Next Domain: Activate the toggle to merge an isolated sample at borehole top with the following domain.
- Merge Isolated Bottom Domain with Previous Domain: Activate the toggle to merge an isolated sample at borehole bottom with the previous domain.
Use ![]() and
and ![]() to add and remove a rule.
to add and remove a rule.
Select an Included Domain then select the Surrounding Domain and a Minimum Length. Included Domain intervals smaller than the Minimum Length will be replaced by the Surrounding Domain. There are two default Surrounding Domains:
- Any: The Included Domain will be replaced by the most likely domain (above or below domain).
- Any (Twice the same domain): The Included Domain will be replaced by the Surrounding Domain if the surrounding domain is the same above and below.
Note: The Smoothing operation is performed before the Grouping operation in the algorithm. If some domains have been grouped together, the Smoothing operation rules should be applied on all the initial domains and not only on the grouped ones.
Parameter Set
Press ![]() to create a new Parameter Set and then modify the parameters of your choice.
to create a new Parameter Set and then modify the parameters of your choice.
Press Compute to generate new Parameter Set’s graphical output. Use the combo box to select a previous computed Parameter Set.
Press Next to get the Clustering Results of the current Parameter Sets.