REBLOCK Process
To access this command:
-
Model ribbon >>Manipulate >> Reblock.
- View the Find Command screen, select REBLOCK and click Run.
- Enter "REBLOCK" into the Command Line and press <ENTER>.
See this process in the Command Table.
Process Overview
Note: This is a superprocess and running it may have an effect on other Datamine files in the project.
Note: This process supports retrieval criteria.
This command re-blocks a block model by user specification of new X, Y and Z parent cell sizes. The origin of the model remains unchanged.
Reblocking can be useful prior to carrying out strategic planning because it can be used to set the model's parent cell size to be representative of the selective mining unit (SMU) size that is being considered.
The user must specify new X, Y and Z parent cell sizes. The number of cells in X, Y and Z (The NX, NY and NZ values) will change if the specified output size is different from the size in the input model. If either the X, Y or Z parent cell sizes are set to zero or are undefined the size in the input model will be retained.
By default, when calculating values in the new output cells, values for all fields are weighted by mass using either the specified DENSITY field or DENSITY parameter value. Alternative field treatments can be used by specifying fields to be 1) Volume Weighted 2) Dominant 3) Additive 4) Selected according to minimum value and 5) Selected according to maximum value.
The new cells sizes can be any size and do not have to be an integer multiple of the input model parent cell size. Care should be taken if re-blocking to a smaller cells size because very large models can be specified. An error message is displayed if the selected cell sizes would cause the output model to exceed the system limits .
All numeric fields in the input model are copied to the output file. Alpha fields defined as Dominant will also be copied to the output file. However any undefined alpha fields will not be copied.
The REBLOCK process can be considered to be an enhanced REGMOD process. To regularize a block model (i.e. remove sub cells) the @FULLCELL parameter can be set to 1, the X, Y and Z sizes do not need to be defined.
Input Model File
The input MODEL file is a normal or rotated block model that can contain parent cells and subcells.
Note: By default, when calculating the output cell values, all numeric fields are weighted by mass using the DENSITY field and DENSITY parameter. Optionally fields can be treated using alternative methods: volume weighted, dominant, additive, minimum or maximum.
*DENSITY Field
The numeric *DENSITY field is used to identify the density values in the input block model file. If values are absent then the @DENSITY parameter value is used. The density values are used for weighting values by mass. The default field name is DENSITY.
FILLVOL and VOIDVOL Fields
When only parent cells are being created in the output model (by setting @FULLCELL=1) the output model will also contain a *FILLVOL field that contains the proportion of each parent cell that was filled by cells in the input model. The default name is FILLVOL. If the specified *FILLVOL field exists in the input model and only parent cells are being generated then the values for this field will be replaced in the output model.
The *VOIDVOL field contains the proportion of the cell in the output model that was not filled by cells in the input model. Therefore VOIDVOL + FILLVOL = 1. values for VOIDVOL and FILLVOL are between 0 and 1.
If @FULLCELL=1 and @UNMODGRD=1 or 2 then any unmodelled volume will be assigned default grades and will be combined with the modelled cells to make a full parent cell. In this case the FILLVOL field defines the proportion of the OUT parent cell that comes from cells and subcells in the IN model and the VOIDVOL field defines the proportion of the OUT parent cell that was unmodelled in the IN model.
Volume Weighted Fields
By default values in the output model are calculated by weighting them against mass. However up to fifteen fields can optionally be volume weighted by specifying their field names using *VFWFLD1 to *VFWFLD15. Volume weighted fields must be numeric.
Dominant Fields
Up to ten fields can be specified to be dominant using *DOMFLD1 to *DOMFLD10. Dominant fields can be numeric or alpha. By default the dominant value is determined by comparing the volume of each value in the new output reblocked cells, the value with the maximum volume will be selected. Dominant fields are useful for attributes such as rock types or fields where the value needs to be copied from the input to the output model. To set the dominant value by mass set the @DBYMASS parameter to 1.
Additive Fields
Up to ten fields can be specified to be additive using *ADDFLD1 to *ADDFLD10. When input model cells are split the value per unit volume is used to calculate the total in the new output parent cells. For example, if two input cells each with an additive value of 50 are being combined into a new reblocked cell, the new cell will have a value of 100. Additive fields must be numeric. Additive fields are useful for fields such as revenue and cost.
If a field with absent data values is defined as additive and @ABSGRADE=2 then the absent values will be replaced by the default value defined in the Data Definition of the IN file. This must be expressed in terms of value per unit volume.
Also if a field is additive and @FULLCELL=1 and @UNMODGRD=1 or 2 then any unmodeled volume will be assigned default grades from the Data Definition. This must be expressed in terms of value per unit volume.
Minimum and Maximum Fields
Up to five fields each can be specified to be minimum or maximum using *MINFLD1 to *MINFLD5 or *MAXFLD1 to *MAXFLD5 respectively. No volume or mass comparisons are done. The minimum (or maximum) value contained in the collection of cells that comprise the output parent cell is copied to the output cell.
Parameter Specifications
XINC, YINC and ZINC Parameters
The @XINC, @YINC and @ZINC parameters are used to specify the parent cell size in the output model. They can be any size and do not have to be an integer multiple of the input model parent cell size dimensions. The values can be smaller or larger than the input models sizes. The origin of the output model is the same as the input model. The number of cells in X, Y and Z is adjusted in the output model to make the extents of the output model approximately the same as the input model.
FULLCELL Parameter
If the @FULLCELL parameter is set to one then every cell in the output model is a parent cell and the model will contain the extra fields *FILLVOL and *VOIDVOL. These fields contain the proportion, between zero and 1, of the output parent cell that is completely filled by cells in the input model.
=0: the output model will only include cells and subcells that existed in the input model
=1: the output model will contain just parent cells. FILLVOL and VOIDVOL fields will be created to show the proportion of modelled and unmodelled cells
DBYMASS Parameter
Set to 1 to determine all dominant field values by mass rather than volume. The default is zero, to determine dominant values by volume.
=0 : Determine dominant values by volume. This is the default
=1 : Determine dominant values by mass.
DENSITY Parameter
The @DENSITY parameter specifies the density value to be used for mass calculations if no DENSITY field exist in the input model or if the DENSITY values in the input model are absent. The @DENSITY parameter is only applied if the @SETABSNT parameter is set to 1. The default value is 1.
SETABSNT Parameter
Set to 1 in order to set absent data density values to the default value defined by the @DENSITY parameter.
=0: do not reset absent density values and report an error
=1: set absent density values to @DENSITY. This is the default
ABSGRADE Parameter
The ABSGRADE parameter controls what action is taken if there are absent data grade values in the input model.
=0: numeric fields with absent data values ( - ) or alpha fields with absent data values (blank) will be reported and the process will terminate. This is the default.
=1: absent data numeric values will be set to zero and absent data alpha values will be set to _ABS
=2: numeric and alpha absent data values will be set to their default values defined in the Data Description of the input model. If the numeric default is absent data then zero will be used. If the alpha default is absent data then _ABS will be used.
UNMODGRD Parameter
The UNMODGRD parameter controls the method used to assign the grade if @FULLCELL=1 but the parent cell includes unmodelled volume ie if FILLVOL<1.
=0: assign the modelled grade values to the full parent cell. This means the unmodelled grades are assumed to be the same as the modelled grades with the parent cell. This is the default.
=1: set all numeric grades of unmodelled volumes to zero and alpha fields to _UNM.
=2: set all grades of unmodelled volumes to their default values defined in the Data Definition of the input model. If the numeric default is absent (-) then a value of zero is used. If the alpha default is absent (blank) then _UNM is used.
In all cases the FILLVOL and VOIDVOL fields are based on the modelled cells only. If the default value of an additive field is applied to the unmodelled volume then it must be defined per unit volume.
The density of unmodelled volumes is defined using the @UNMODDEN parameter
UNMODDEN Parameter
The UNMODDEN parameter defines the density of unmodelled volumes. It is only used if @FULLCELL=1 and @UNMODGRD=1 or 2. The default value is 1.
Input Files
|
Name |
Description |
I/O Status |
Required |
Type |
|
MODIN |
Input block model file |
Input |
Yes |
Block Model |
Output Files
|
Name |
I/O Status |
Required |
Type |
Description |
|
MODOUT |
Output |
Yes |
Block Model |
Reblocked output model file |
Fields
|
Name |
Description |
Source |
Required |
Type |
Default |
|
DENSITY |
Density field. If the model does not include a DENSITY field then the density can be set with parameter @DENSITY. The output file will always contain a DENSITY field. |
IN |
Yes |
Numeric |
Undefined |
|
FILLVOL |
The proportion of each full cell in the output model that was filled with cells in the input model. Only output if @FULLCELL=1. |
IN |
No |
Numeric |
Undefined |
|
VOIDVOL |
The proportion of each full cell in the output model that was filled with cells in the input model. Calculated as 1 - FILLVOL. Only output if @FULLCELL=1. |
IN |
No |
Numeric |
Undefined |
|
VWFLD1-15 |
Field(s) to be volume weighted |
IN |
No |
Numeric |
Undefined |
|
DOMFLD1-10 |
Field(s) to be treated as dominant. |
IN |
No |
Numeric |
Undefined |
|
ADDFLD1-10 |
Field(s) to be treated as additive |
IN |
No |
Numeric |
Undefined |
|
MINFLD1-5 |
Field(s) to be set using minimum value |
IN |
No |
Numeric |
Undefined |
|
MAXFLD1-5 |
Field(s) to be set using maximum value |
IN |
No |
Numeric |
Undefined |
Parameters
|
Name |
Description |
Required |
Default |
Range |
Values |
|
XINC |
New X parent cell size |
No |
20 |
2,+ |
Undefined |
|
YINC |
New Y parent cell size |
No |
20 |
0,+ |
Undefined |
|
ZINC |
New Z parent cell size |
No |
20 |
Undefined |
Undefined |
|
FULLCELL |
Set to 1 to output a model containing only parent cells |
No |
0 |
0,1 |
0,1 |
|
DBYMASS |
Set to 1 to determine dominant values by mass rather than volume. The default is zero, to determine dominant values by volume. |
No |
0 |
0,1 |
0,1 |
|
DENSITY |
Grade to be applied as a topcut, if TOPCUT is set to 1. Any value greater than TOPGRADE will be reset to equal to TOPGRADE. |
No |
1 |
0,1 |
0,1 |
|
SETABSNT |
Set to 1 to set any absent density values to the default density using the @DENSITY value. |
No |
0 |
0,1 |
0,1 |
|
ABSGRADE |
The ABSGRADE parameter controls what action is taken if there are absent data grade values in the input model. |
No |
0 |
0,2 |
0,1,2 |
|
UNMODGRD |
The UNMODGRD parameter controls the method used to assign the grade if @FULLCELL=1 but the parent cell in the OUT model includes unmodelled volume ie if FILLVOL is less than 1. The default is (0). |
No |
0 |
0,2 |
0,1,2 |
|
UNMODDEN |
The UNMODDEN parameter defines the density of unmodelled volumes. It is only used if @FULLCELL=1 and @UNMODGRD=1 or 2. The default value is 1. |
No |
1 |
Undefined |
Undefined |
Examples
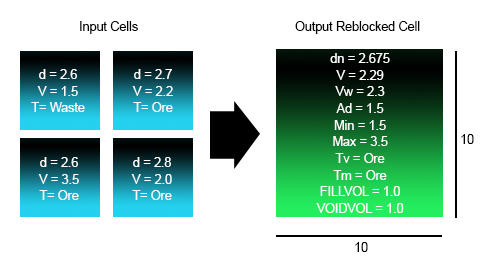
Reblocking Example 1
In the following example, the input model is 5*5*1 and the output model is 10*10*1. FULLCELL =1.
|
Density (Vol Weight): |
dn=(2.6*25+2.7*25+2.6*25+2.8*25)/(25+25+25+25) = 2.675 |
| Mass Weighted V: |
V = (2.6*1.5*25+2.7*2.2*25+2.6*3.5*25+2.8*2.0*25)/(2.6*25+2.7*25+2.6*25+2.8*25) = 2.29 |
| Volume Weighted V: |
Vw = (1.5*2+2.2*25+3.5*25+2.0*25)/(25+25+25+25) = 2.3 |
| Additive V: |
Ad = 25*(1.5/25)+25*(2.2/25)+25*(3.5/25)*2.0/25)=9.2 |
| Minimum V: |
Min=1.5 |
| Maximum V: |
Max=3.5 |
| Dominant Tv: |
T = max(25*waste,25*2.7*Ore+25*2.6*Ore+25*2.8*Ore) = Ore |
| Dominant Tm: |
T = max(25*2.6*Waste,25*2.7+Ore+25*2.6*Ore+25*2.8*Ore) = Ore |
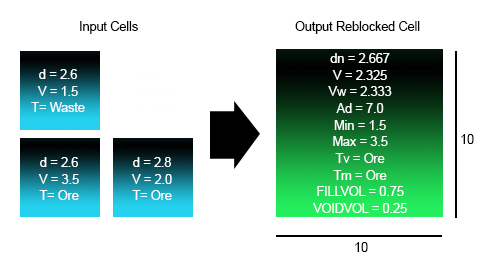
Reblocking Example 2
In the following example, the input model is 5*5*1 and the output model is 10*10*1. FULLCELL =1.
|
Density (Vol Weight): |
dn = (2.6*25 + 2.6*25 + 2.8*25) / (25+25+25) = 2.667 |
| Mass Weighted V: | V = (2.6*1.5*25 + 2.6*3.5*25 + 2.8*2.0*25) / (2.6*25 + 2.6*25 + 2.8*25) = 2.325 |
| Volume Weighted V: | Vw = (1.5*25 + 3.5*25 + 2.0*25) / (25 + 25 + 25) = 2.333 |
| Additive V: | Ad = 25*(1.5/25) + 25*(3.5/25) + 25*(2.0/25) = 7.0 |
| Minimum V: | Min=1.5 |
| Maximum V: | Max=3.5 |
| Dominant Tv: | T = max (25*Waste, 25*Ore + 25*Ore) = Ore |
| Dominant Tm: | T = max (2.6*25*Waste, 25*2.6*Ore + 25*2.8*Ore) = Ore |
Resource Reporting
Note that in these examples the grade values are for a volume of 10*10*1*FILLVOL. Therefore, if process TONGRAD is used to report the resource of the output model then the OREFRAC field must be set to FILLVOL.